Posted by Gregory J Block MSc PhD on Feb 6, 2015
Dr. Robert Bradley is a young faculty member at the Hutch in Seattle. He's particularly savvy at computational biology - as evidenced by his background in mathematics and physics, and the fact that his laptop is always full of green-font gibberish like something out of the Matrix. He and his team have been putting their computational skills to use, and have identified a fundamental process that goes awry when DUX4 is present in cells.
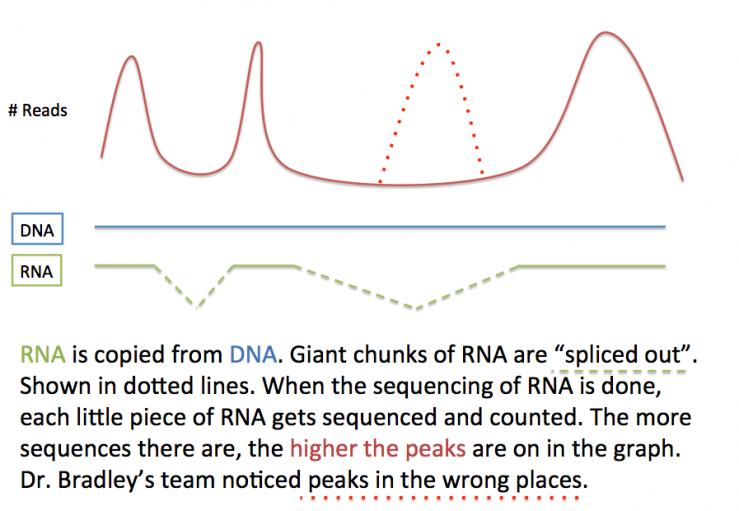
But Dr. Bradley noticed something not-quite-right. When he looked at the peaks and valleys of RNAs from thousands of genes, they weren’t where they were supposed to be - and it wasn't just because they were being mis-spliced through a process called "alternative splicing". Something else was going on.
One way our ingenious bodies survey for mis-spliced RNAs is through a process called nonsense-mediated decay, or NMD. NMD is a very simple quality control mechanism, like the cell version of a guy at a chocolate factory who picks out all the bad chocolates. NMD ensures that there are no premature stop codons in the final RNA. A stop codon is what tells the protein-making machine to stop making a protein. If you didn’t have stop codons, proteins would never end, like a knitter who doesn’t count their stitches. If an RNA contains a premature stop codon, then it is rapidly destroyed by NMD.
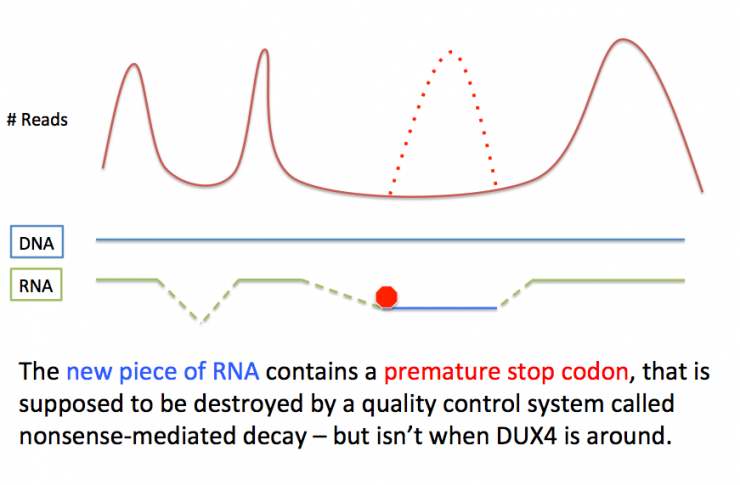
These are very important findings for the field for a number of reasons. First, it highlights that the changes made to cells by DUX4 are global in nature. The data continue to support the central role of DUX4 in muscle pathology. But there is still much to learn about how DUX4 induced NMD in the first place. Once those puzzle pieces are put in place, we’ll begin to have a very clear idea of what DUX4 is doing, and may uncover new targets for the development of therapeutics.
Friends of FSH Research helped support this study through funds from Dr. Tapscott’s grant summarized here. The first author of the paper is Qing Feng, a graduate student in Molecular and Cellular Biology at the University of Washington.
Feng Q, Snider L, Jagannathan S, Tawil R, van der Maarel SM, Tapscott SJ, Bradley RK (2015). A feedback loop between nonsense-mediated decay and the retrogene DUX4 in facioscapulohumeral muscular dystrophy. eLife 10.7554/eLife.04996.
See also: F1000 recommendation.





Connect with us on social media