Posted by George Shaw on Nov 14, 2016
See: Investigation of DUX4 activity during development of hESC-derived skeletal muscle
This project aimed to develop a screening assay using Dr. Dan Miller’s DUX4-reporter with Genea Biocells’ FSHD and unaffected human Embryonic Stem Cell (hESC) lines using the Genea Biocells three stage myogenesis-in-a-dish protocol. One control unaffected and three FSHD hESC lines were transduced with GFP, BFP, or luciferase versions of a DUX4-activated lentiviral reporter and selected using G418 resistance. hESCs were expanded, characterized, banked and DUX4-responsiveness was demonstrated by transfection of GFP reporter lines with a plasmid encoding DUX4.
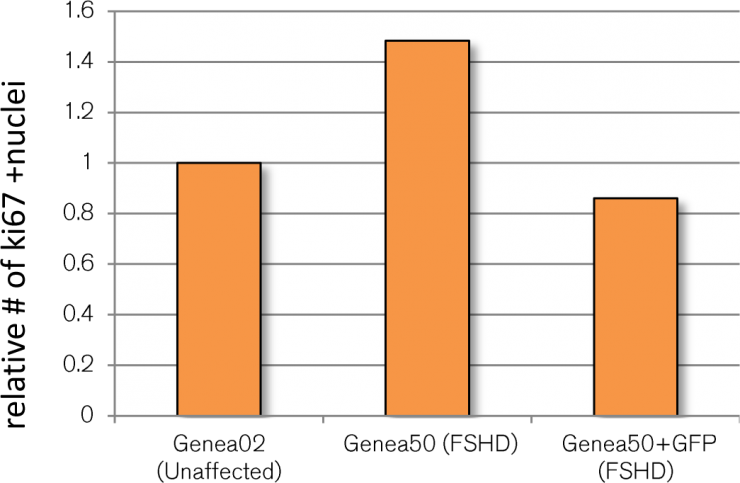
Caron et al. (2016) described an increased FSHD-specific proliferation phenotype during myogenic differentiation as measured by higher levels of ki67+ labelled FSHD cells relative to controls. Since ki67+ is a potentially screenable phenotype and could serve as an independent secondary assay, FSHD and control GFP reporter myotube cultures were also stained for ki67 expression, imaged, and quantified (Figure 1). Although Caron et al reported FSHD-affected Genea50 myotubes to have a larger ki67+ cell population than control Gen02 myotubes, this difference was not seen in cells that had been transduced with the lentiviral reporter. It is worth noting that in order to generate the DUX4-GFP reporter lines, the FSHD and control hESC lines had to undergo a stressful selection process (G418 treatment) followed by extensive expansion and passaging prior to banking. Although these cell banks were fully characterized for pluripotency and their ability to differentiate towards skeletal muscles, the selection process or additional passaging/expansions may have impacted the proliferative phenotype observed in the original lines. Regrettably, these findings rendered the ki67-related FSHD phenotype unsuitable for FSHD therapeutics screening and was abandoned.
Initial observations of the DUX4-GFP reporter at different stages of skeletal muscle differentiation indicated rare reporter activation during Stage 1 (myogenic precursor) followed by a more substantial signal in Stage 3 (myotube). However, the transient and sporadic nature of the DUX4-GFP signal at either stage made immunofluorescence-based quantification inaccurate and difficult. Live cell imaging is a more comprehensive, informative, and accurate quantification approach, especially in situations where activation events are rare. Whereas the differential fusion of myotubes can introduce variability in population-based reporters, such as the DUX4-luciferase construct, long-term BFP reporter live cell imaging requires continuous UV exposure that leads to DNA damage and cytotoxicity in healthy cells. To that end, the luciferase and BFP reporter constructs were abandoned and all experiments described henceforth are based on DUX4-GFP reporter constructs using live-cell quantification. During the course of this study, the Genea Biocells team tested three independent live cell imaging instruments (Cytation 5 by BioTek, Incucyte by Essen Bioscience, JuLi Stage by NanoEntek) and identified the Incucyte as the most reliable and accurate method for high-throughput fluorescent analysis.
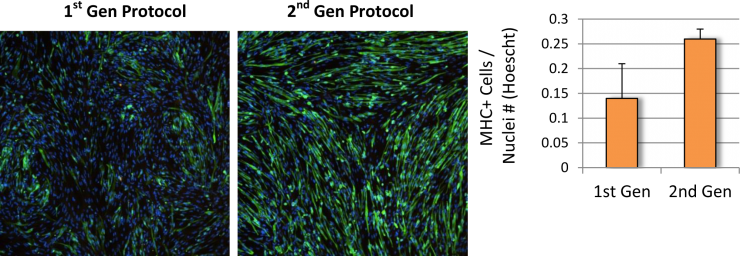
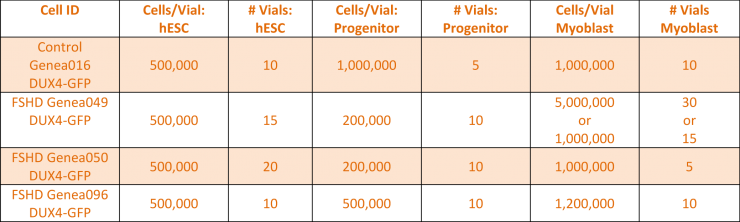
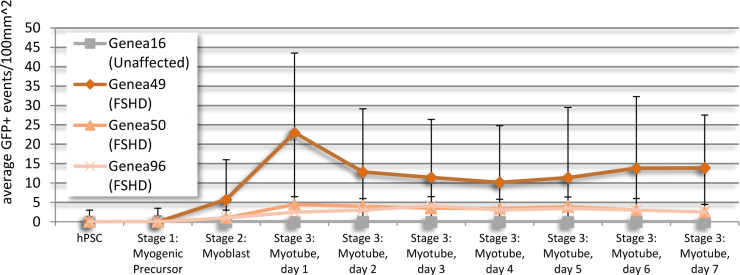
Baseline levels of DUX4 expression were established during the myogenic differentiation of Genea FSHD-affected hESCs DUX4-GFP reporter lines using the newly established live cell imaging instrumentation. These initial experiments were run with the 1st generation Genea Biocells myogenic differentiation protocol and showed very low or undetectable levels of DUX4-reporter expression. Independently, a 2nd generation skeletal muscle differentiation was developed with more consistent and improved yields, as detectable by increased Myosin Heavy Chain + myotubes (Figure 2), the cell type previously reported to express DUX4. To improve the DUX4-GFP detection levels, significant efforts were invested to generate and bank new stocks of hESC DUX4-GFP reporter lines, Stage 1 Precursors and Stage 2 Myoblasts (Table 1) using the 2nd generation method. Using the newly derived lines, all hESC-GFP reporter lines were continuously imaged during all stages of the myogenic differentiation and DUX4-GFP activation levels were quantified (Figure 3). GFP-activation events were rarely detected in the Stage 1 FSHD myogenic precursor stage. DUX4-GFP expression increased through the Stage 2 myoblast stage and reached a maximal and sustained levels in Stage 3 myotubes. Genea016 control, unaffected cells did not activate DUX4-GFP to detectable levels in any stage and Genea049-GFP was identified as the highest DUX4-activating FSHD line. Even at peak DUX4 expression levels, activation events were infrequent, with the maximal reading of 43 DUX4-GFP activation events per 300,000 cells assayed in Genea049-GFP myotubes (.014% GFP+ cells). To increase the probability of capturing DUX4-GFP activity, subsequent experiments were carried out using FSHD Genea049-GFP myoblasts derived using the 2nd generation differentiation method.
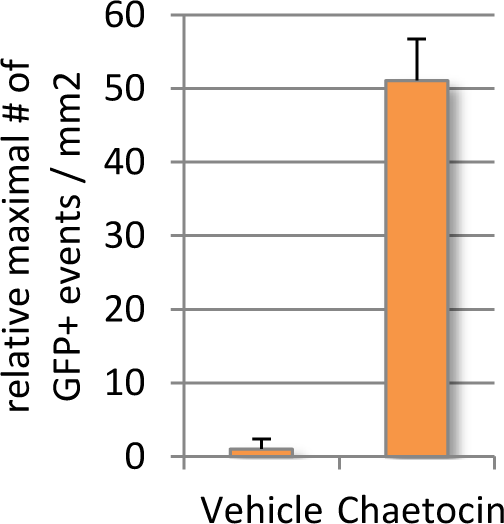
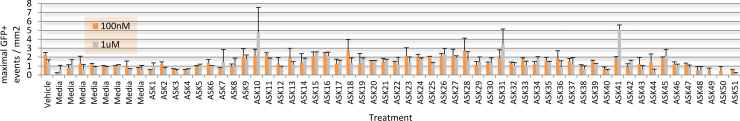
Given that endogenous DUX4 expression in Genea FSHD-affected myogenic cells is rare and difficult to detect in a high-throughput manner, Gen049-GFP myoblast cells were treated with published compounds reported to exacerbate DUX4 expression (Zeng et al., 2009). Treatment with Chaetocin, a DUX4-activating histone methyltransferase inhibitor, resulted in 12x higher DUX4-GFP activation than that seen in the vehicle control treated cells (Figure 4). The concentrations of Chaetocin required for observation of DUX4-GFP activation were toxic to both FSHD and control, healthy myoblasts beginning 40h after exposure. In order to identify conditions that increase DUX4 activation with minimal drug-induced toxicity, a library of 51 proprietary small molecules was assembled, targeting epigenetic, gene regulation, and FSHD-specific pathways. Gen049-GFP myoblasts were treated with 1uM or 100nM of each compound for 24 hours then differentiated to myotubes, and live cell imaging for DUX4-GFP reporter fluorescence was performed on all wells (with outside edges of plates excluded to reduce variability). This screen identified ASK10, ASK31, and ASK41 as novel alternative DUX4-activating small molecules (Figure 5).
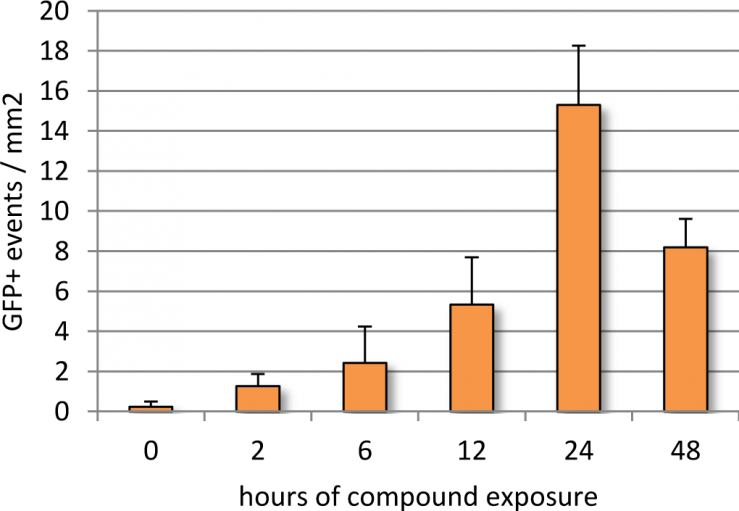
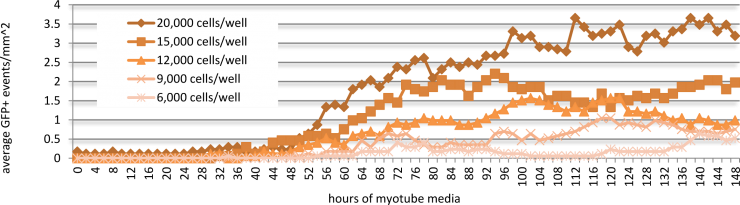
Follow-up experiments were performed with ASK10 in order to optimize exposure for maximal DUX4-GFP activation. In a time-course study, Genea049-GFP myoblasts were exposed to 1uM of ASK10 for 0, 2, 6, 12, 24, and 48 hours in Stage 2, prior to myotube induction (Figure 6). 24 hours of compound treatment induced the highest DUX4-GFP expression, whereas 48 hours of exposure resulted in noticeable drug-induced cell death and therefore lower detectable events. The cell plating density of Genea049-GFP myoblasts before exposure to ASK10 also altered the detectable levels of DUX4-GFP by live cell imaging analysis (Figure 7). The highest plating density of cells (20,000 cells per well or 62,500 cells per cm2) yielded the greatest DUX4-GFP activation. In future studies, more concentrations of cells will be tested to further optimize density dependent DUX4-GFP activation. Overall, the discovery of novel DUX4-activating compounds not only generated a scalable and quantifiable hESC-derived chemical model of FSHD, but also helped identify potential nodes for therapeutic intervention for this devastating disease.
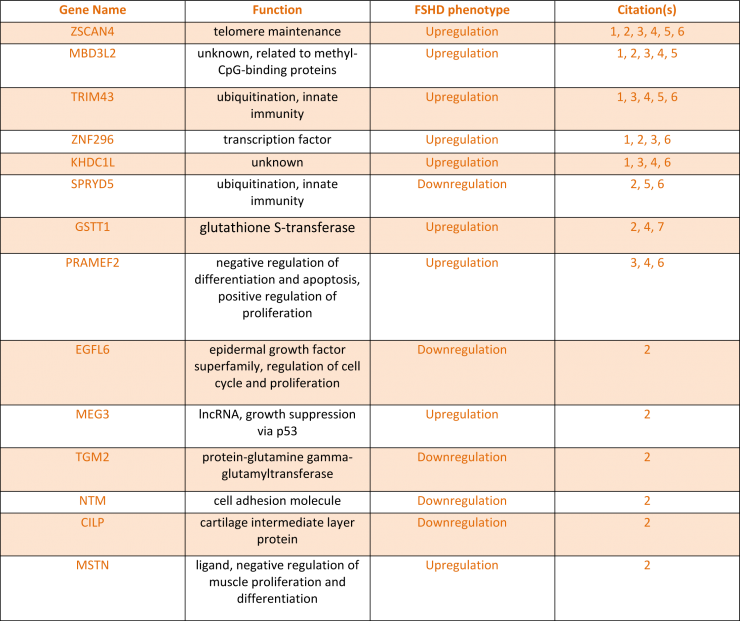
A panel of 14 genes whose expression level change has been documented as an FSHD phenotype was compiled from extensive literature searches of FSHD and DUX4-induced transcript quantification assays (Table 2). Now that conditions have been optimized for both Genea myotube formation and DUX4 activation using molecules targeting different regulatory pathways, we will apply the TruSeq method for gene expression analysis to measure how FSHD-associated gene changes are altered in FSHD and upon DUX4 activation in hESC-derived myogenic cells.
To date, efforts towards the development of a high-throughput, scalable FSHD myogenesis model for therapeutic screening have successfully yielded many important insights. This project’s scope yielded the establishment of a sensitive and quantifiable live cell imaging platform for DUX4 detection, adaptation of the reporter lines to the second generation muscle differentiation protocol, validation of reported chemically induced DUX4 expression, and identification of novel small molecule modulators of DUX4. Armed with these, we will continue our endeavors in FSHD therapeutic research by focusing on secondary assay development and testing of a TruSeq panel to monitor DUX4 target gene expression in FSHD and unaffected stem cell derived skeletal muscle cultures.





Connect with us on social media